More Information
Submitted: 13 August 2019 | Approved: 15 August 2019 | Published: 16 August 2019
How to cite this article: Efebera YA, Ruppert AS, Ngankeu A, Garman S, Kumchala P, et al. Serum MicroRNA-155 in Acute Graft-Versus-Host-Disease (aGVHD). Int J Bone Marrow Res. 2019; 2: 079-082.
DOI: 10.29328/journal.ijbmr.1001007
Copyright License: © 2019 Efebera YA, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Keywords: Mir-155; Graft versus host disease, MicroRNA; Allogeneic stem cell transplant
Serum MicroRNA-155 in Acute Graft-Versus-Host-Disease (aGVHD)
Yvonne A Efebera1*, Amy S Ruppert1, Apollinaire Ngankeu1, Sabrina Garman1, Prasanthi Kumchala1, Alan Howard2, Steven M Devine1, Parvathi Ranganathan1 and Ramiro Garzon1
1Division of Hematology, Department of Internal Medicine and Comprehensive Cancer Center, The Ohio State University, Columbus, Ohio, USA
2Blood and Marrow Transplant Clinical Trial Network, National Marrow Donor Program, USA
*Address for Correspondence: Yvonne A Efebera, Associate Professor, Division of Hematology, Department of Internal Medicine and Comprehensive Cancer Center, The Ohio State University, Columbus, Ohio, USA, Tel: 614-293-2268; Fax: 614-293-7526; Email: [email protected]
Allogeneic hematopoietic stem cell transplant (alloHSCT) is a curative treatment for many hematologic malignancies. Unfortunately, about 30-50% of all recipients undergoing alloHSCT develop acute graft-versus-host-disease (aGVHD), which is associated with high morbidity and mortality [1,2]. Treatment of aGVHD involves the use of immune suppressive drugs such as high dose of steroids that leads to further immunosuppression and risk for opportunistic infections. Often patients are refractory to steroids therapy making the prognosis dismal. Thus, it is critical to identify robust biomarkers to detect aGVHD before onset of clinical symptoms so that therapeutic strategies can be implemented that may result in better treatment responses and less toxicity.
Over the past few years, several plasma proteins have been identified and validated to predict aGVHD occurrence, severity and treatment response. These plasma biomarkers include among others Interleukin 2 receptor, tumor necrosis alpha receptor 1, interleukin 8, hepatocyte growth factor and receptor suppression of tumorigenicity 2 (ST2) [3-7]. These biomarkers are now recognized as robust and reliable by experts in the field [8,9]. However, protein-based assays such as ELISA and mass spectrometry have drawbacks such as the need for high volume and high affinity quality antibodies with the required affinity and specificity for the target [9]. Further, focusing only on proteins may not be enough to develop powerful and comprehensive biomarkers. Thus, it will be important to investigate whether cell free DNA or RNA that are detectable in the plasma or serum predict aGVHD and provide prognostic information and whether these biomarkers may complement the current validated protein-based tests.
In the past years, circulating microRNAs (miRs) have emerged as possible candidates for aGVHD biomarkers. A panel of four miRs (miR-423, miR-199a-3p, miR-93*, and miR-377) were found to be over-expressed in the plasma of aGVHD patients 16 days before diagnosis as compared to non-GVHD patients [10]. One study showed that plasma miR-586 was upregulated in patients with aGVHD [11], and 2 independent studies identified miR-155 to be upregulated in the serum [12] and whole blood [13] of patients that developed aGVHD versus those who did not. Since miR-155 expression in T cells and in antigen presenting cells (APCs) is critical for aGVHD pathogenesis [14,15], we sought to validate the previous reports on high levels of circulating miR-155 in aGVHD. Thus, to investigate whether serum miR-155 was elevated in patients at time of aGVHD onset following alloHSCT as compared to matched controls, we measured miR-155 expression from serum samples obtained from patients enrolled in the Blood and Marrow Transplant Clinical Trial Network (BMT CTN) 0101 study, a randomized, double-blind trial of fluconazole vs. voriconazole for prevention of invasive fungal infection after alloHSCT [16]. From 98 patients enrolled on this trial, samples at aGVHD onset within 100 days from alloHSCT were matched with controls without aGVHD (n=49 pairs). Patient samples were matched on day from alloHSCT (+/- 7 days in 96% of pairs), histology of disease (acute leukemia and MDS vs. CML vs. lymphoma), receipt of GVHD prophylaxis, and type of donor (related vs. unrelated). The samples were obtained from the National Heart, Lung, and Blood Institute Biologic Specimen Repository (NHLBI Biorepository).
Serum total RNA was extracted using the miRNeasy Serum/Plasma Kit (Cat. no.217184, QIAGEN, Valencia, CA, USA) with a modified protocol. 200 µL of serum sample was carefully transferred to a new 2-mL DNA-LoBind tube (Eppendorf, Hamburg, Germany). Serum was mixed with 1 ml QIAzol Lysis Reagent. The mixture was placed on a vortex mixer at 3000 rpm for 30 s and then left at room temperature for 5 min to allow complete inactivation of serum RNases. 4 µL of a mixture of serial dilution (1, 0.1 and 0.01 pg/ µL) of pooled synthetic microRNA was spiked into the homogenate lysis mixture. From here, the manufacturer’s protocols were followed for RNA extraction (Cat. no.217184, QIAGEN, Valencia, CA, USA). Qiagen miScript PCR System was used for reverse transcription and RT-qPCR. 100ng of RNA was converted into cDNA using the miScript II Reverse Transcription Kit (Cat. N.218161) and with the HiSpec Buffer according to the manufacturer’s protocol. The RT-qPCR was performed with the miScript SYBR® Green PCR Kit (Cat. N. 218073) in a total volume of 20μl per reaction containing 2μl diluted cDNA according to the manufacturer’s protocol. C-elegans microRNAs spike-in control expression was used for normalization. The miScript universal primer and the miRNA-specific miScript primer assay for Hs_miR-155 (Cat. MS00031486), Ce_miR-238 (Cat. MS00019439), Ce_miR-54 (Cat. MS00019894) and Ce_miR-39 (Cat. 219610) (all from QIAGEN), were used to detect mature miRNA. Individual real-time PCR assays were performed in triplicate wells on an ABI 7500 real-time PCR system (Applied Biosystems) and results were averaged. A paired t-test was used to compare -∆CT values for serum miR-155 expression between aGVHD patients and non-aGVHD controls. Conditional logistic regression was used to model the probability of being called the case within a matched pair as a function of miR-expression and important or confounding variables not accounted for by the matching. All tests were two-sided and statistical significance was declared for p<0.05.
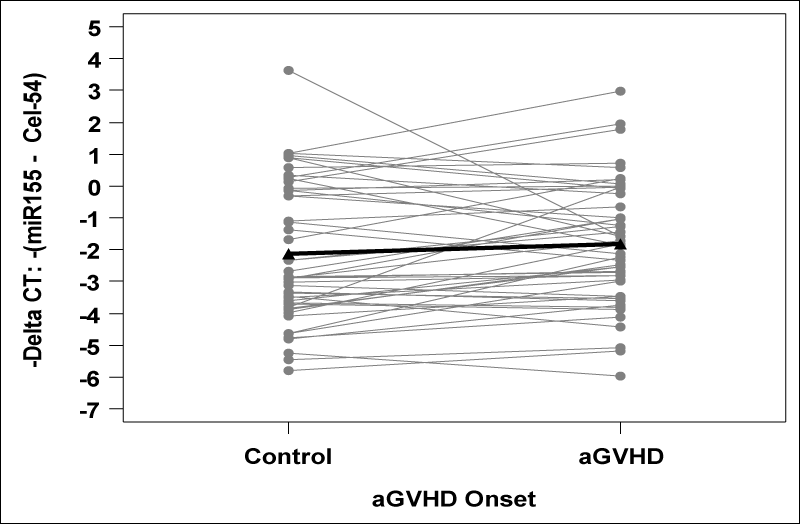
Patient characteristics are provided in table 1. Among the 49 cases, 39 had grade 2 aGVHD and 10 had grade 3 aGVHD at onset. The median time to aGVHD onset was 21 days from transplant (range: 4 – 91). We found no significant difference in miR-155 expression between aGVHD cases and matched controls (p=0.16; Figure 1). Although expression was higher in 31 cases (63%), the average -∆CT was higher by 0.29 (95% CI: -0.12 to 0.70) and corresponded to a non-significant relative fold change of 1.22 (95% CI: 0.92 to 1.63). The only clinical variable significantly associated with aGVHD was donor source, with 84% of cases receiving stem cell source from peripheral blood compared to 61% of controls (p=0.02; Table 1). When adjusting for donor source, the association between miR-155 and presence of aGVHD remained non-significant (p=0.16).
| Table 1: Patient Characteristics | ||||
| Characteristic | All Patients n=98 |
Cases n=49 |
Controls n=49 |
p-value* |
| Patientage in years at transplant, no. (%) <30 31-50 >50 |
27 (28) 49 (50) 22 (22) |
12 (24) 23 (47) 14 (29) |
15 (31) 26 (53) 8 (16) |
0.30 |
| Patient sex, no. (%) Male Female |
49 (50) 49 (50) |
22 (45) 27 (55) |
27 (55) 22 (45) |
0.32 |
| Patient race, no. (%) White Non-white Unknown |
87 (92) 8 (8) 3 |
44 (90) 5 (10) 0 |
43 (93) 3 (7) 3 |
0.42 |
| Diseasediagnosis, no. (%) Acute Leukemia/MDS CML Lymphoma |
78 (80) 12 (12) 8 (8) |
39(80) 6 (12) 4 (8) |
39 (80) 6 (12) 4 (8) |
NA |
| Performancescore, no. (%) 100 90 <80 |
32 (33) 56 (57) 10 (10) |
15 (31) 30 (61) 4(8) |
17 (35) 26 (53) 6 (12) |
0.66 |
| Prophylaxistreatment, no. (%) Fluconazole Voriconazole |
48 (49) 50 (51) |
24 (49) 25 (51) |
24 (49) 25 (51) |
NA |
| HLAscore, no. (%) 5/6 6/6 |
2 (2) 96 (98) |
1 (2) 48 (98) |
1 (2) 48 (98) |
1.00 |
| Stemcell source Marrow PBSC |
27 (28) 71 (72) |
8 (16) 41 (84) |
19(39) 30 (61) |
0.02 |
| Donorrelationship, no. (%) Related Unrelated |
64 (65) 34 (35) |
32 (65) 17 (35) |
32 (65) 17 (35) |
NA |
| Abbreviations: MDS=myelodysplastic syndrome; CML=chronic myelogenousleukemia; PBSC=peripheral blood stem cell; NA=not applicable; P-values fromunivariable conditional logistic regression models that account for theindividual matching of cases to controls based on disease diagnosis,prophylaxis treatment, and donor relationship are presented. Models cannot befit for the variables upon which the matching was performed and thus p-valuesare not applicable. | ||||
Figure 1: miR-155 expression is depicted as -∆CT values, with higher values corresponding to higher expression. Using a paired t-test, the -∆CT was on average 0.29 units higher (95% CI: -0.12 to 0.70) in cases compared to matched controls (p = 0.16). This corresponds to a non-significant relative fold change of 1.22 (95% CI: 0.92 to 1.63) in cases versus matched controls..
Hence, no significant difference in miR-155 expression was observed in the serum of patients at onset of aGVHD compared to those who did not have aGVHD. This is in contrast to previous reports showing an upregulation of serum and whole blood miR-155 in aGVHD [12,13]. Possible reasons for this discrepancy could be 1: Sample size and patient population: The sample size of the previous studies that assessed mir-155 association with aGVHD are small (N=25-28) [12,13]. While our study is also small, it is almost twice as large and had at least 80% power to detect a relative fold change in miR-155 expression of 1.54 when the standard deviation of the differences in -∆CT values between cases and controls was as large as 1.5, as was observed in our dataset. The patient population studied in the previous studies were wide, including non-hematologic disease, while this study focused on hematologic malignancies with 92% being leukemias and myelodysplasia. 2: Timing of assessment of samples: The clinical trial from which patient samples were obtained was conducted between 2003 and 2006 and samples were stored in the NHLBI biorepository. It is therefore possible that some degradation may have occurred. It is however noteworthy that there is little overlap in the circulating miRNAs identified as prognostic by profiling studies, which could be explained by the different sources used (plasma vs. serum), profiling methods and small numbers of patients. Despite these findings, this in no way rules out a role for microRNAs in the pathogenesis of acute GVHD, but at the moment they do not serve as reliable biomarkers. We realize the limitations of this study: lack of samples from peak aGVHD and lack of serial samples that precludes evaluation of miR-155 at a time point prior to aGVHD onset or change in miR-155 expression as a prognostic factor. There is therefore the need to conduct comprehensive studies with serial samples obtained from a sufficiently large number of patients with robust profiling methods to derive reliable RNA or DNA circulating biomarkers.
Financial Support and Grant for this study
National Heart Lung and Blood Institute (NHLBI) R21
This manuscript was prepared using materials from BMT CTN clinical trial 0101 obtained from the NHLBI Biologic Specimen and Data Repository Information Coordinating Center. Support for the BMT CTN 0101 trial was provided by grant U10HL069294 to the Blood and Marrow Transplant Clinical Trials Network from the National Heart, Lung, and Blood Institute, the National Cancer Institute along with contributions by Pfizer Inc. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funders.
YE is the co-recipient of the NHLBI-R21, and wrote the Letter; AR is the statistician and also edited the letter; AN and PR did the analysis in the lab and edited the letter; SG and PR are the research coordinators in the lab; AH is with the NMDP that coordinated sample release, and edited the letter; SMD did major edit of the Letter; RG is senior author, recipient of the NHLBI-R21 and did Major editing of the letter.
- Blazar BR, Murphy WJ, Abedi M. Advances in graft-versus-host disease biology and therapy. Nat Rev Immunol. 2012; 12: 443-458 PubMed: https://www.ncbi.nlm.nih.gov/pubmed/22576252
- Ferrara JL, Levine JE, Reddy P, Holler E. Graft-versus-host disease. Lancet. 2009; 373: 1550-1561. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/19282026
- Paczesny S. Discovery and validation of graft-versus-host disease biomarkers. Blood. 2013; 121: 585-594. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/23165480
- Paczesny S, Braun TM, Levine JE, Hogan J, Crawford J, et al. Elafin is a biomarker of graft-versus-host disease of the skin. Sci Transl Med. 2010; 2: 13ra2. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/20371463
- Paczesny S, Krijanovski OI, Braun TM, Choi SW, Clouthier SG, et al. A biomarker panel for acute graft-versus-host disease. Blood. 2009; 113: 273-278. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/18832652
- Reichenbach DK, Schwarze V, Matta BM, Tkachev V, Lieberknecht E, et al. The IL-33/ST2 axis augments effector T-cell responses during acute GVHD. Blood. 2015; 125: 3183-3192. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/25814531
- Vander Lugt MT, Braun TM, Hanash S, Ritz J, Ho VT, et al. ST2 as a marker for risk of therapy-resistant graft-versus-host disease and death. N Engl J Med. 2013; 369: 529-539. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/23924003
- Ali AM, DiPersio JF, Schroeder MA. The Role of Biomarkers in the Diagnosis and Risk Stratification of Acute Graft-versus-Host Disease: A Systematic Review. Biol Blood Marrow Transplant. 2016; 22: 1552-1564. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/27158050
- Paczesny S, Levine JE, Braun TM, Ferrara JL. Plasma biomarkers in graft-versus-host disease: a new era? Biol Blood Marrow Transplant. 2009; 15: 33-38. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/19147075
- Xiao B, Wang Y, Li W, Baker M, Guo J, et al. Plasma microRNA signature as a noninvasive biomarker for acute graft-versus-host disease. Blood. 2013; 122: 3365-3375. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/24041574
- Wang Y, Zhao X, Ye X, Luo H, Zhao T, et al. Plasma microRNA-586 is a new biomarker for acute graft-versus-host disease. Ann Hematol. 2015; 94: 1505-1514. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/26051902
- Xie LN, Zhou F, Liu XM, Fang Y, Yu Z, et al. Serum microRNA155 is increased in patients with acute graft-versus-host disease. Clin Transplant. 2014; 28: 314-323. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/24494749
- Atarod S, Ahmed MM, Lendrem C, Pearce KF, Cope W, et al. miR-146a and miR-155 Expression Levels in Acute Graft-Versus-Host Disease Incidence. Front Immunol. 2016; 7: 56. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/27014257
- Chen S, Smith BA, Iype J, Prestipino A, Pfeifer D, et al. MicroRNA-155-deficient dendritic cells cause less severe GVHD through reduced migration and defective inflammasome activation. Blood. 2015; 126: 103-112. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/25972159
- Ranganathan P, Heaphy CE, Costinean S, Stauffer N, Na C, et al. Regulation of acute graft-versus-host disease by microRNA-155. Blood. 2012; 119: 4786-4797. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/22408260
- Wingard JR, Carter SL, Walsh TJ, Kurtzberg J, Small TN, et al. Randomized, double-blind trial of fluconazole versus voriconazole for prevention of invasive fungal infection after allogeneic hematopoietic cell transplantation. Blood. 2010; 116: 5111-5118. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/20826719